Multivariate time series models¶
Multivariate time series models are designed to capture the dynamic of multiple time series simultaneously and leverage dependencies across these series for more reliable predictions.
The basic requirements in order to use VAR are:
- You need atleast two time series (variables)
- The time series should influence each other.
The vector autoregressive VAR(p) model extends the AR(p) model to k series by creating a system of k equations where each contains p lagged values of all k series. Multivariate time series models allow for lagged values of other time series to affect the target. This effect applies to all series, resulting in complex interactions.
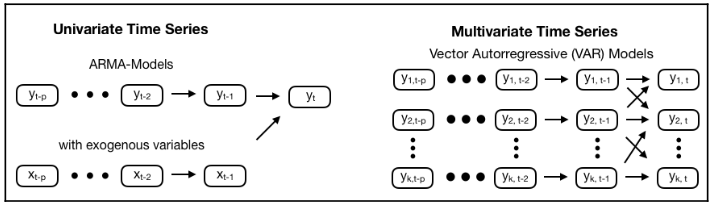
In the VAR model, each variable is modeled as a linear combination of past values of itself and the past values of other variables in the system. Since you have multiple time series that influence each other, it is modeled as a system of equations with one equation per variable (time series). A VAR(1) model for k=2 takes the following form:
Since the Y terms in the equations are interrelated, the Y’s are considered as endogenous variables, rather than as exogenous predictors.
VAR(p)models also require stationarity.
How to use the VAR model for macro fundamentals forecasts¶
Imports and Settings¶
import os
import sys
import warnings
from datetime import date
import pandas as pd
import pandas_datareader.data as web
import numpy as np
from numpy.linalg import LinAlgError
from numpy import cumsum, log, polyfit, sqrt, std, subtract
import matplotlib.pyplot as plt
import matplotlib.transforms as mtransforms
import matplotlib.cm as cm
import seaborn as sns
from scipy.stats import spearmanr, pearsonr
from scipy.stats import probplot, moment
import statsmodels.api as sm
import statsmodels.tsa.api as smt
from statsmodels.tsa.api import VAR, VARMAX
from statsmodels.tsa.stattools import acf, q_stat, adfuller
from statsmodels.graphics.tsaplots import plot_acf, plot_pacf
from statsmodels.tsa.stattools import grangercausalitytests
from statsmodels.tsa.stattools import adfuller
from statsmodels.stats.stattools import durbin_watson
from sklearn.metrics import mean_squared_error, mean_absolute_error
%matplotlib inline
warnings.filterwarnings('ignore')
sns.set(style='darkgrid', context='notebook', color_codes=True)
Helper Functions¶
Hurst Exponent¶
def hurst(ts):
lags = range(2, 100)
tau = [sqrt(std(subtract(ts[lag:], ts[:-lag]))) for lag in lags]
poly = np.polyfit(log(lags), log(tau), 1)
return poly[0]*2.0
Correlogram Plot¶
def plot_correlogram(x, lags=None, title=None):
lags = min(10, int(len(x)/5)) if lags is None else lags
fig, axes = plt.subplots(nrows=2, ncols=2, figsize=(12, 8))
x.plot(ax=axes[0][0])
q_p = np.max(q_stat(acf(x, nlags=lags), len(x))[1])
stats = f'Q-Stat: {np.max(q_p):>8.2f}\nADF: {adfuller(x)[1]:>11.2f} \nHurst: {round(hurst(x.values),2)}'
axes[0][0].text(x=.02, y=.85, s=stats, transform=axes[0][0].transAxes)
probplot(x, plot=axes[0][1])
mean, var, skew, kurtosis = moment(x, moment=[1, 2, 3, 4])
s = f'Mean: {mean:>12.2f}\nSD: {np.sqrt(var):>16.2f}\nSkew: {skew:12.2f}\nKurtosis:{kurtosis:9.2f}'
axes[0][1].text(x=.02, y=.75, s=s, transform=axes[0][1].transAxes)
plot_acf(x=x, lags=lags, zero=False, ax=axes[1][0])
plot_pacf(x, lags=lags, zero=False, ax=axes[1][1])
axes[1][0].set_xlabel('Lag')
axes[1][1].set_xlabel('Lag')
fig.suptitle(title, fontsize=20)
fig.tight_layout()
fig.subplots_adjust(top=.9)
Unit Root Test¶
def test_unit_root(df):
return df.apply(lambda x: f'{pd.Series(adfuller(x)).iloc[1]:.2%}').to_frame('p-value')
Durbin Watson Test¶
def durbin_watson_test(df=None, resid=None):
cols, stat = [], []
out = durbin_watson(resid)
for col, val in zip(df.columns, out):
cols.append(col)
stat.append(round(val, 2))
dw_test = pd.DataFrame(stat, index=cols)
return dw_test
Load Data¶
We will use monthly data on industrial production and, consumer sentiment provided by the Federal Reserve's data service. We will use the familiar pandas-datareader library to retrieve data from 1972 through 2020:
sent = 'UMCSENT'
df = web.DataReader(['IPGMFN', 'UMCSENT'], 'fred', '1972', '2020-03').dropna()
df.columns = ['ip', 'con_sent']
df.info()
Plot Series¶
df.plot(subplots=True, figsize=(13,13));
EDA¶
g = sns.clustermap(df.corr(), annot=True)
ax = g.ax_heatmap
bottom, top = ax.get_ylim()
ax.set_ylim(bottom + 0.5, top - 0.5)
plt.title('Pairwise Correlations')
plt.show()
sns.jointplot(x=df.ip.pct_change().dropna(), y=df.con_sent.pct_change().dropna(), kind="reg");
The next step in the analysis is to check for causality amongst these series. The Granger’s Causality test and the Cointegration test can help us with that.
Testing Causation using Granger’s Causality Test¶
The Granger causality test is a statistical hypothesis test for determining whether one time series is useful in forecasting another. Ordinarily, regressions reflect "mere" correlations, but Clive Granger argued that causality in economics could be tested for by measuring the ability to predict the future values of a time series using prior values of another time series. Since the question of "true causality" is deeply philosophical, and because of the post hoc fallacy of assuming that one thing preceding another can be used as a proof of causation, econometricians assert that the Granger test finds only "predictive causality". Rather than testing whether Y causes X, the Granger causality tests whether Y forecasts X.
A time series X is said to Granger-cause Y if it can be shown, usually through a series of t-tests and F-tests on lagged values of X (and with lagged values of Y also included), that those X values provide statistically significant information about future values of Y. The original definition of Granger causality does not account for latent confounding effects and does not capture instantaneous and non-linear causal relationships.
In simpler terms, the past values of time series (X) do not cause the other series (Y). So, if the p-value obtained from the test is lesser than the significance level of 0.05, then, you can safely reject the null hypothesis.
The below code implements the Granger’s Causality test for all possible combinations of each time series in a given dataframe and stores the p-values of each combination in matrix form.
maxlag = 12
test = 'ssr_chi2test'
def grangers_causation_matrix(data, variables, test='ssr_chi2test', verbose=False):
df = pd.DataFrame(np.zeros((len(variables), len(variables))), columns=variables, index=variables)
for c in df.columns:
for r in df.index:
test_result = grangercausalitytests(data[[r, c]], maxlag=maxlag, verbose=False)
p_values = [round(test_result[i+1][0][test][1],4) for i in range(maxlag)]
if verbose: print(f'Y = {r}, X = {c}, P Values = {p_values}')
min_p_value = np.min(p_values)
df.loc[r, c] = min_p_value
df.columns = [var + '_x' for var in variables]
df.index = [var + '_y' for var in variables]
return df
grangers_df = grangers_causation_matrix(df, variables = df.columns)
plt.figure(figsize=(8,5))
sns.heatmap(grangers_df, annot=True)
b, t = plt.ylim()
b += 0.5
t -= 0.5
plt.ylim(b, t)
plt.show()
Find Significant Pairs in a p-value matrix¶
pairs = grangers_df.unstack()
pairs = pairs.sort_values(kind="quicksort")
mask = pairs < 0.05
print("Significan Pairs")
print(pairs[mask])
Looking at the P-Values in the above table, you can pretty much observe that most the variables (time series) in the system are interchangeably causing each other. This makes this system of multi time series a good candidate for using VAR models to forecast.
Cointegration Test¶
Cointegration test helps to establish the presence of a statistically significant connection between two or more time series. Cointegration is a statistical property of two or more time-series variables which indicates if a linear combination of the variables is stationary.
When two or more time series are cointegrated, it means they have a long run, statistically significant relationship.
This is the basic premise on which Vector Autoregression(VAR) models is based on. So it is common to implement the cointegration test before starting to build VAR models.
Implementation of the cointegration test in python’s statsmodels can see below:
def find_cointegrated_pairs(dataframe, critial_level = 0.05):
n = dataframe.shape[1]
pvalue_matrix = np.ones((n, n))
keys = dataframe.columns
pairs = []
for i in range(n):
for j in range(i+1, n):
series1 = dataframe[keys[i]]
series2 = dataframe[keys[j]]
result = sm.tsa.stattools.coint(series1, series2)
pvalue = result[1]
pvalue_matrix[i, j] = pvalue
if pvalue < critial_level:
pairs.append((keys[i], keys[j], pvalue))
return pvalue_matrix, pairs
pvalue_matrix, pairs = find_cointegrated_pairs(df)
coint_pvalue_matrix_df = pd.DataFrame(pvalue_matrix)
g = sns.clustermap(coint_pvalue_matrix_df, xticklabels=df.columns,yticklabels=df.columns, annot=True,
figsize=(8, 8))
plt.title('Cointegration P-value Matrix')
ax = g.ax_heatmap
bottom, top = ax.get_ylim()
ax.set_ylim(bottom + 0.5, top - 0.5)
plt.show()
pairs
Pair is not significant
Check for Stationarity¶
Since the VAR model requires the time series you want to forecast to be stationary, it is customary to check all the time series in the system for stationarity.
A stationary time series is one whose mean and variance does not change over time.
So, how to test for stationarity?
There is a suite of tests called unit-root tests. The popular ones are:
- Augmented Dickey-Fuller Test (ADF Test)
- KPSS test
- Philip-Perron test
Let’s implement the ADF Test by using our plot_correlogram() function which retuns a p-value for the ADF test.
for i in df.columns:
plot_correlogram(df[i], lags=100, title=f'{i}')
Stationarity Transform¶
Log-transforming the industrial production and differencing both series using lag 12 yields stationary results:
df_transformed = pd.DataFrame({'ip': np.log(df.ip).diff(12),
'con_sent': df.con_sent.diff(12)}).dropna()
Inspect Correlograms¶
for i in df_transformed.columns:
plot_correlogram(df_transformed[i], lags=100, title=f'{i}_log_diff')
test_unit_root(df_transformed)
All the series are now stationary.
Split the Series into Training and Testing Data¶
split = int(len(df) * 0.91)
df_train, df_test = df_transformed.iloc[:split], df_transformed.iloc[split:]
print(df_train.info())
print(df_test.info())
How to Select the Order (P) of VAR model¶
To select the right order of the VAR model, we iteratively fit increasing orders of VAR model and pick the order that gives a model with least BIC.
test_results = {}
for p in range(5):
for q in range(5):
if p == 0 and q == 0:
continue
print(f'Testing Order: p = {p}, q = {q}')
convergence_error = stationarity_error = 0
try:
model = VARMAX(df_train, order=(p,q), trend='n')
model_result = model.fit(maxiter=1000, disp=False)
except LinAlgError:
convergence_error += 1
except ValueError:
stationarity_error += 1
print('\nAIC:', model_result.aic)
print('BIC:', model_result.bic)
print('HQIC:', model_result.hqic)
print('------------------------')
test_results[(p, q)] = [model_result.aic,
model_result.bic,
convergence_error,
stationarity_error]
test_results = pd.DataFrame(test_results).T
test_results.columns = ['AIC', 'BIC', 'convergence', 'stationarity']
test_results.index.names = ['p', 'q']
test_results.info()
We aim to minimize BIC:¶
sns.heatmap(test_results.BIC.unstack(), fmt='.2f', annot=True, cmap='Blues_r')
b, t = plt.ylim()
b += 0.5
t -= 0.5
plt.ylim(b, t)
plt.show()
test_results.sort_values('BIC').head()
Estimating with the best VAR Model¶
We will estimate a VAR(1,0) model using the statsmodels VARMAX implementation (which allows for optional exogenous variables) with a no trend using the first 164 observations. The output contains the coefficients for both time series equations.
model = VARMAX(df_train, order=(1,0), trend='n').fit(maxiter=1000)
model.summary()
Plot Diagnostics¶
statsmodels provides diagnostic plots to check whether the residuals meet the white noise assumptions, which are not exactly met in this simple case:
Industrial Production
model.plot_diagnostics(variable=0, figsize=(13,8), lags=24)
plt.gcf().suptitle('Industrial Production - Diagnostics', fontsize=20)
plt.tight_layout()
plt.subplots_adjust(top=.9);
Consumer Sentiment
model.plot_diagnostics(variable=1, figsize=(13,8), lags=24)
plt.gcf().suptitle('Consumer Sentiment - Diagnostics', fontsize=20)
plt.tight_layout()
plt.subplots_adjust(top=.9);
Impulse-Response Function¶
median_change = df_transformed.diff().quantile(.5).tolist()
model.impulse_responses(steps=12, impulse=median_change).plot.bar(subplots=True, figsize=(13,13));
Check for Serial Correlation of Residuals (Errors) using Durbin Watson Statistic¶
If there is any correlation left in the residuals, then, there is some pattern in the time series that is still left to be explained by the model. In that case, the typical course of action is to either increase the order of the model or induce more predictors into the system. Checking for serial correlation is to ensure that the model is sufficiently able to explain the variances and patterns in the time series.
The value of this statistic can vary between 0 and 4. The closer it is to the value 2, then there is no significant serial correlation. The closer to 0, there is a positive serial correlation, and the closer it is to 4 implies negative serial correlation.
durbin_watson_test(df=df_transformed, resid=model.resid)
Generate Predictions¶
Out-of-sample predictions can be generated as follows:
start = 440
preds = model.predict(start=490, end=len(df_transformed)-1)
preds.index = df_transformed.index[490:]
fig, axes = plt.subplots(nrows=2, figsize=(12, 8))
df_transformed.ip.iloc[start:].plot(ax=axes[0], label='actual', title='Industrial Production')
preds.ip.plot(label='predicted', ax=axes[0])
trans = mtransforms.blended_transform_factory(axes[0].transData, axes[0].transAxes)
axes[0].legend()
trans = mtransforms.blended_transform_factory(axes[0].transData, axes[1].transAxes)
df_transformed.con_sent.iloc[start:].plot(ax=axes[1], label='actual', title='Sentiment')
preds.con_sent.plot(label='predicted', ax=axes[1])
fig.tight_layout();
Out-of-sample forecasts¶
A visualization of actual and predicted values shows how the prediction lags the actual values and does not capture non-linear out-of-sample patterns well:
forecast = model.forecast(steps=24)
fig, axes = plt.subplots(nrows=2, figsize=(12, 8))
df_transformed.ip.plot(ax=axes[0], label='actual', title='Liquor')
preds.ip.plot(label='predicted', ax=axes[0])
axes[0].legend()
df_transformed.con_sent.plot(ax=axes[1], label='actual', title='Sentiment')
preds.con_sent.plot(label='predicted', ax=axes[1])
axes[1]
fig.tight_layout();
Model evaluation mean absolute error MAE¶
mean_absolute_error(forecast, df_transformed.iloc[-24:])